What is NanoString Technology?
For using the infrastructure please 👉 CLICK HERE to download the form
Please send the completed requisition form to arnab@nii.ac.in
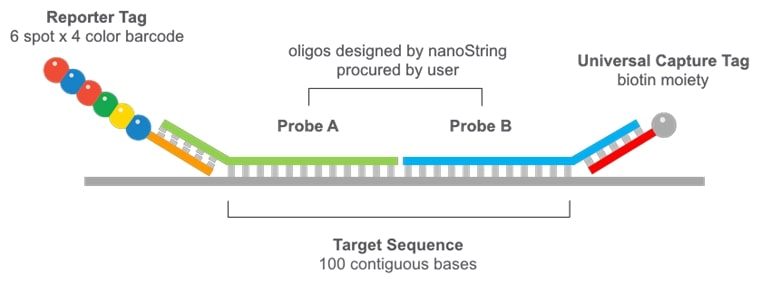
NanoString is a new and very different technology from any other available today. It is essentially a single-molecule counting system. It works by attaching molecular barcodes to target molecules of interest by nucleic acid base-pairing, a process that is simple, robust and well-understood. The molecular barcodes consist of a series of 6 fluorescent "spots", each of which can be one of 4 colours. The different combinations of ordered spot colours create different "barcodes" that uniquely identify each molecule in an experiment. In this way, several million molecules can be counted from a sample. Experimentally, you can use up to 800 different barcodes to detect and very precisely quantify target molecules, providing great scope for multiplexing. You can detect any number of targets up to the full 800-plex from a single sample, and without changing the effort required, the time is taken or the experimental protocol in any way.
Hallmarks of the Technology
There are several hallmarks of nanoString technology that set it apart:
1. Automation
Molecule processing (cleanup and arraying onto a glass surface) and counting (using high-content screening, or automated microscopy) is automated and identical for all applications, including DNA, RNA, miRNA, proteins and combination thereof. It is thus routine, simple to implement with only minutes of hands-on time and does not require a dedicated operator.
2. Enzyme-free robustness
NanoString is an enzyme-free technology. For gene expression applications this means there is no reverse transcription (RT) or PCR steps. Only simple base-pairing in a hybridisation reaction is used to process samples, removing the most troublesome variables and providing robustness: Assays are immune to PCR inhibitors, degradation and sample impurity issues that plague the PCR and technologies that use it (such as RNAseq and digital PCR). Note that some exotic nanoString applications harness enzymatically-driven sample pre-hybridisation processing, such as pre-enrichment PCR cycles, endonuclease digestion or ligation.
3. Digital Precision
Counting molecules is an automated process that is completely objective and done without any (subjective) human involvement. There are no intensity signals, the nCounter instrument sees a perfect barcode or it does not. Anything else is ignored (including imperfect barcodes which are treated correctly as random events). The benefits of a purely digital signal cannot be overstated; it provides a simplicity and precision that is unprecedented and makes even the most complex experiments reproducible between operators and laboratories.
4. No need for technical replicates
Although this sounds confronting, given how essential technical replicates are to most technologies (in particular the PCR), it is true for two reasons. Firstly, there is no ambiguity regarding assay dropout events. Dropouts are common with PCR-based assays and require technical replicates to identify them. With nanoString, all analytes (including all the many controls built into every assay) are equally affected since they are all parallel-processed in the same tube and thus a dropout is instantly recognised. Secondly, because the system works by population sampling, once enough molecules are counted the precision is high and easily calculated. A technical replicate is effectively just more molecule counts and is thus redundant. Peer review referees know this and nowadays you no longer see technical replicates in publications. The focus is directed at informative biological replicates instead.
5. Unprecedented Sensitivity
NanoString assays detect molecules with zeptomole (10-21) sensitivity. The limit of detection (LOD ) is ~500 molecules and the limit of quantification is (LOQ) ~1000 molecules. Although we think of PCR as the gold standard for sensitivity, this is not usually the case compared to nanoString. The LOQ for the PCR is ~100 molecules (from many publications, e.g Gasparic et al 2008. Experimentally, however, triplicate assays are usually run, taking the number of molecules required up to 300 per target. Further, PCR is not a multiplex technology (in actual use), so the number of molecules required is ~300 per target. Nowadays we realise that for many applications (such as gene expression analysis) several references (housekeeping) genes are required along with additional contextural targets. Since samples analysed by nanoString are not split and technical replicates are not required, even a simple experiment of 2 targets + 3 housekeeping genes requires 1500 molecules (2× 300 + 3× 300) if analysed using RT-PCR.To extrapolate, an 800-target RT-PCR experiment requires 240,000 molecules (800×3×100) and a vast amount of precision quantitative sample splitting into different tubes, where nanoString does the same experiment in a single tube using the same sample input for any plex up to 800. This is especially important when samples are rare or a limited resource.
What Molecules Can Be Counted?
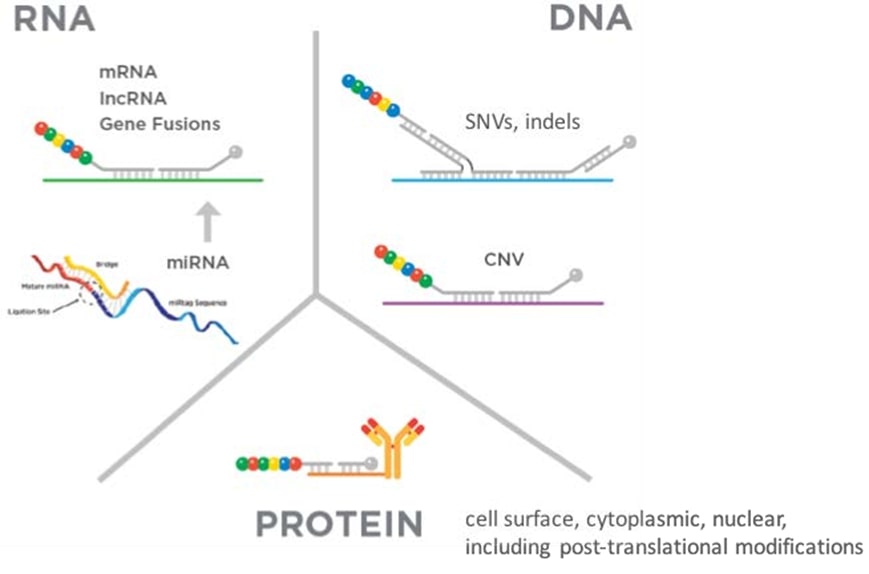
Several new chemistries have recently been introduced that allow the system to detect and quantify three broad analyte classes: DNA, RNA and proteins. The three molecule classes can be counted from a single sample either separately or simultaneously, enabling what nanoString calls "3D Biology".
Although an entirely new paradigm, 3D Biology "super signatures" are quickly gaining momentum in the translational space as the information content from each analyte class is not additive, but acts with a multiplier effect. Further, the workflow is simple and the data is easy to harmonise as it is all captured as simple digital (numerical) molecule counts.
Built for Translational Applications
NanoString's origin, in the Leroy Hood laboratories at the Institute for Systems Biology in Seattle WA, was aimed at the clinical and translational application from inception.
NanoString Technologies Inc. are very proactive in the translational space, with support for the full range of applications from discovery science through to clinical use. Highlights include:
1. Commercialisation of human diagnostics
Clinical-grade assay development, analytical and clinical validation and full commercialisation of the Prosigna® Breast Cancer Prognostic Gene Signature Assay. Prosigna is FDA510(k) cleared and CE-marked for in vitro diagnostic use in the United States and EU. It is now TGA-approved in Australia and is about to launch in New Zealand and other countries. Largely because the assay uses nanoString technology, Prosigna has been shown to outperform all other competing commercial assays.
2. Pharma collaboration and partnerships
NanoString recently signed the three largest companion diagnostics agreements in history, with over 20 more in the pipeline. (Press releases for agreements with Merck, HalioDx, Mepation and Astellas and Cellgene) and recently announced the availability of the PDL-1 checkpoint inhibitor companion diagnostic known as TIS (Tumour Inflammation Signature) as part of the IO 360 Panel product in order to promote translational applications more broadly.
3. Shown to be superior for clinical-grade applications
NanoString was recently the subject of an independent, detailed and peer-reviewed technology review by Astra-Zeneca (UK) titled Evaluating Robustness and Sensitivity of the NanoString Technologies nCounter Platform to Enable Multiplexed Gene Expression Analysis of Clinical Samples. The conclusions were: “We found the practical aspect of using the nCounter platform favourable over any other technique used with minimal time needed and reduced margin for user error due to few handling steps during the preparation of reaction” and “This study demonstrates the nCounter technology offers key advantages, including sensitivity, technical reproducibility, and robustness for analysis of FFPE samples and strong evidence of the utility of the nCounter for mRNA gene expression analysis in clinical tissue".
4. Direct Translational Collaborations
NanoString is serious about translating assays into clinical and diagnostic use. They operate a Translational Research Program (TRP) with the express goal of research collaborations leading to continuous improvement and expansion of products leading to clinical development. There are numerous partnerships around the world between hospitals and nanoString for the development of clinical-grade assays for diagnostics and personalised medicine (such as at Roswell Park Cancer Institute in New York, Brigham and Women's Hospital in Boston, the Hospital for Sick Children (SickKids) in Toronto (where they are developing gene fusion panel tests for sarcomas, they see nanoString is their "next gold standard" and are exploring its use more broadly, in fact, “anywhere you'd use a PCR assay".
5. Clinical-grade chemistry specifically for Laboratory Developed Tests (LDTs)
Nanostring developed and released Elements™ chemistry specifically to support the simple implementation and rapid translation of assays into LDTs. Elements chemistry uses a unique“open architecture” chemistry with performance benefits (extremely low background counts) and the ability to define and create any unique assay using only two pools of assay-specific oligos coupled with standardised off-the-shelf FDA-registered General Purpose Reagents (GPRs).
Chemistry : -
NanoString projects can be broadly pided into projects using either
A) Regular Chemistry, which interrogates many targets (up to 800) from few samples, or
B) PlexSet™ Chemistry, which interrogates fewer targets (up to 96) from many samples
A: Regular Chemistry
Assay costs vary depending on:
1. The number of targets in the assay, which amortises costs over more or fewer targets, and
2. The number of assays runs, which for custom assays will determine the price of custom codeSets (there are economies of scale with larger synthesis scales)
B: PlexSet™ Chemistry
PlexSet chemistry is a new option that uses 2-dimensional multiplexing to condense more samples into a single instrument run for applications where throughput is paramount. A nCounter instrument processes 12 assays in a run but with PlexSet each assay position processes 8 separate pooled samples for 96 samples in total. With PlexSet, you can choose to multiplex up to 12- or 24-plex (with 48- and 96-plex options available later in 2017). The 2nd plex dimension uses different optical barcode sets (TagSets) to differentiate up to 8 pooled samples in each instrument flowcell lane (so the barcode can identify not only the target gene but which sample it came from in the pool). PlexSet uses the following Elements-based chemistry, where the Probe A and Probe B oligos define each target in an assay. Pooled Probe A and Probe B oligos define a particular assay.
Hidden costs: Plasticware Usage Comparison of PlexSet vs RT-PCR
Assay Design Support
All nanoString assays are designed by a bioinformatics team within nanoString dedicated to the task. Assay design is a free service with no financial commitment required. It is a consultative process between the customer and nanoString and, depending on the complexity of the project brief, may take from 24 hours to several weeks to complete, with customer input along the way as required.
Assays can be designed for any target, from any species. Where publicly available, nanoString uses the latest sequence databases for up-to-date designs. Every possible design permutation is tested for best assay performance, targeting the maximum number of transcripts by default (although this can be tailored to meet the design brief). A design is not completed until the customer is completely happy with it. Each design includes a report detailing the sequences targeted and other relevant details, including any pertinent notes such as which pseudogenes could not be avoided etc. In the case of oligo-only PlexSet and Elements chemistry designs, an IDT order form is provided already populated with oligo sequences and ready to submit to the oligo manufacturer.
A separate free online tool nDesign™ Gateway, allows the researcher to quickly find targets of interest by pathway or function and identify which Panel products already contain those genes.
Data Analysis Support
NanoString provides a free data analysis tool called nSolver™. nSolver is free and is essentially a wizard-based front-end query engine for the Microsoft R statistical computing platform. nSolver runs on both Mac and PCs and is supported by comprehensive documentation, online video training and a dedicated email support line (support@nSolver.com). There are also options for full-project analysis as a separate paid service direct from nanoString for very complex projects or where bioinformatics support is required but not available to the researcher. nSolver is at version 4 and quite sophisticated despite its simple wizard interface. It is now more powerful than many of the high-end statistical packages available that typically cost many thousands of dollars per inpidual license.
Comparison to PCR and Other Technologies
NanoString compares to other technologies according to best-in-class principles: Concordance with existing technologies is always high and better than when other technologies are compared amongst themselves. This is because nanoString is the least variable, pure digital technology. There are many published examples of this and a few key publications are tabulated below.
| Paper | Title | Reproducibility | PlatformValidation | vs qPCR | vs other | FFPE | Ge ne Sig | CNV | miRNA | Notes |
|---|---|---|---|---|---|---|---|---|---|---|
| Northcott 2012 | Rapid, reliable, and reproducible molecular sub-grouping of clinical medulloblastoma samples | ✔ | ✔ | ✔ | Reproducibility, FFPE, GeneSignature, Standard to Elements, SVM ClassPrediction, Why NS is superior to Microarray and IHC | |||||
| Scott 2014 | Determining cell-of-origin subtypes of diffuse large B-cell lymphoma using gene expression informalin-fixed paraffin-embedded tissue | ✔ | ✔ | FFPE, Gene Signature Development, CDx, why NS is superior to IHC | ||||||
| Scott 2015 | Prognostic Significance of Diffuse Large B-Cell Lymphoma Cell of Origin determined by Digital Gene Expression in Formalin-Fixed Paraffin-Embedded Tissue Biopsies | ✔ | ✔ | A lot to lot variability, Reproducibility, Low tumour content (10%), FFPE and Gene Signature | ||||||
| Veldman-Jones 2014 | Reproducible, quantitative and flexible molecular sub-typing of clinical DLBCL samples using the nanoString nCounter system | ✔ | ✔ | IHC, Affy | ✔ | Platform validation, FFPE, Gene Signature, Reproducibility, vs Affy, IHC, qNPA, Astra Zeneca | ||||
| Lira 2013 | Multiplexed Gene Expression and Fusion Transcript Analysis to Detect ALK Fusions in Lung Cancer | ✔ | ✔ | Gene Fusion, FFPE (500ngRNA), Gene Signature, Tumourcontent 10%, Seq and RT- PCR validation, Alternative to FISH and IHC, Cost Effective, Pfizer | ||||||
| Lira 2014 | A Single-Tube Multiplexed Assay for Detecting ALK, ROS1 and RET Fusions in Lung Cancer | ✔ | ✔ | |||||||
| Nielson 2014 | Analytical validation of the PAM50-based Prosigna Breast Cancer Prognostic Gene Signature Assay and nCounter Analysis System using formalin-fixed paraffin-embedded breast tumour specimens | ✔ | ✔ | ✔ | ✔ | Key validation, FFPE, Gene Signature, Reproducibility | ||||
| Russell 2013 | Quantitation of Gene Expression in Formaldehyde-Fixed and Fluorescence-Activated Sorted Cells | ✔ | ✔ | The best proof of our strength in FFPE | ||||||
| Tan IB 2015 | High-depth sequencing of over 750 genes supports the linear progression of primary tumours and metastases in most patients with liver-limited metastatic colorectal cancer | ✔ | IHC, FISH | ✔ | CNV, FFPE, CNV validation by IHC, FISH or qPCR | |||||
| Teruel-Montoya 2014 | Micro RNA Expression Differences in Human Hematopoietic Cell Lineages Enable Regulated Transgene Expression | ✔ | ✔ | Working with blood cells, blood cell purification, data analysis, housekeeping miRNA selection, qPCR validation, Blood miRNA | ||||||
| Cascione 2013 | Integrated MicroRNA and mRNA Signatures Associated with Survival in Triple Negative Breast Cancer | ✔ | ✔ | mRNA-miRNA Integration both from NS, FFPE (1995- 2005), data analysis, discovery biology using panels | ||||||
| Barczak 2012 | RNA Signatures allow rapid identification of pathogens and antibiotic susceptibilities | Pathogen Detection, antibiotic resistance, other species, lysates (RLT buffer) and TRIzo l |
Some Notes on RT-PCR
The PCR is coming under increasing scrutiny due to its inherently high variability, the amount of validation required, improper use of controls and reference (housekeeping) genes in most applications and generally poor implementation in many laboratories around the world. Published examples include:
Bustin & Nolan 2017: Talking the talk, but not walking the walk: RT-qPCR as a paradigm for the lack of reproducibility in molecular research
This is the strongest attack on the PCR I've yet seen and it is an indication of the change in attitudes to the PCR of late. Some quotes:
“The ostensible simplicity of RT-qPCR conceals a complexity that extends to every step of the workflow", “We conclude that the majority of published RT-qPCR data are likely to represent technical noise", “Unwillingness to take remedial action in general and for RT-qPCR, in particular, means that much taxpayers’ money and donors’ generosity continues to be wasted."
Bustin et al 2014: Variability of the Reverse Transcription Step: Practical Implications
The reverse transcription step is routinely overlooked but contributes an enormous bias and variability to gene expression experiments. This up-to-date appraisal of the RT step shows that the problem is rife and affects any method that uses an RT step (which includes RNAseq). Quote: "Results showed variability was sufficiently large to call into question the validity of many published data that rely on quantification of cDNA".
Dijkstra et al: Critical appraisal of quantitative PCR results in colorectal cancer research: Can we rely on published qPCR results?
This paper shows how poorly PCR is practised by assessing all the qPCR colorectal cancer publications from 2006 to August 2013 for the number of reference genes used and whether they had been validated. It is quite horrifying. In short:
- The validity of 97% (173/179) of the publications could not be assessed
- 92% of publications use only a single reference gene, and only 6% use more than two
- 70% of RT-qPCR publications use just one of three reference genes (ACTB/ GAPDH/18S
- 13% of publications report whether the chosen reference gene was validated
Lanoix et al: Quantitative PCR Pitfalls: The Case of the Human Placenta
The conclusion (which should be no surprise) is that if you choose the wrong reference gene you will get completely opposite results. Using a single reference gene is simply invalid.
Drozd et al: Doxorubicin Treatment of Cancer Cells Impairs Reverse Transcription and Affects the Interpretation of RT-qPCR Results.
The real message is to use a panel of reference genes and validate them (and also follow the MIQUE Guidelines scrupulously).
Maximally Orthogonal for Validation Use
NanoString is the ideal validation technology since it is the most orthogonal here: