बिचित्र क. बिस्वाल
My laboratory aims at understanding the structural, functional and essentiality aspects of proteins of metabolic pathways using structural biology, biochemistry and in vivo studies and utilizes the outcome of these basic research studies for drug discovery for infectious diseases. We majorly focus on designing anti-TB molecules primarily through a target-based approach.
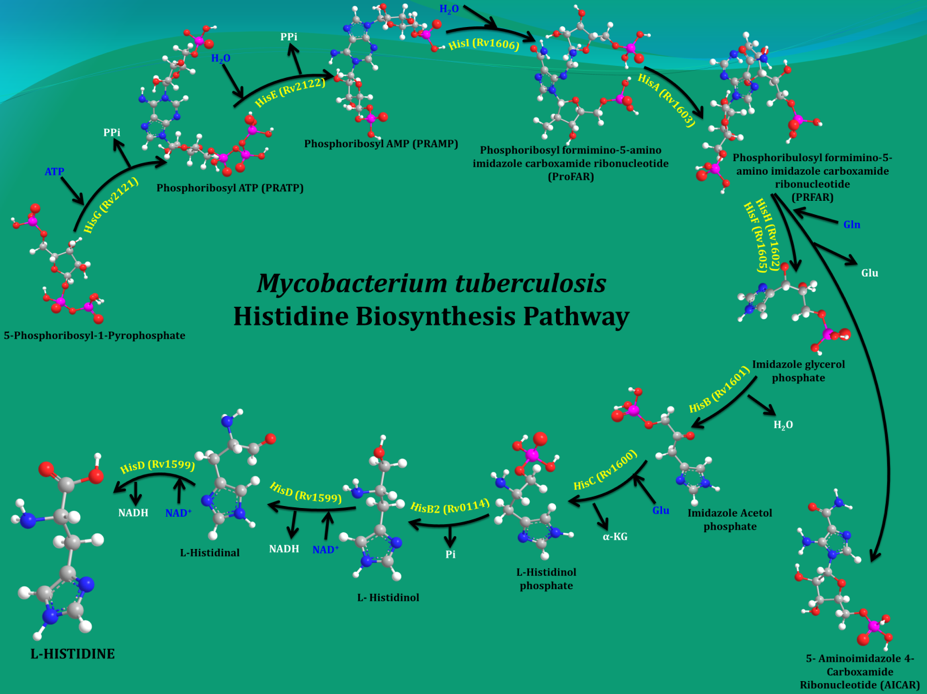
Bacteria, plants and fungi all have enzymes to synthesize histidine (His). However, it cannot be synthesized de novo in humans due to the absence of equivalent enzymes. Mycobacterium tuberculosis (Mtb), the organism that causes tuberculosis (TB) in humans, makes His in 10 steps by employing 11 enzymes (Figure 1). Earlier studies by other groups and current data from my own group showed that even a single His pathway gene knockout significantly attenuates the bacterial load than the wild strain. In light of this result and the fact that His constitutes one of the building blocks of protein synthesis, inhibition of the enzymes that are involved in its biosynthesis gleams a rational strategy for new anti-TB agents design. In this context, the 3-D structures of these enzymes will be helpful in elucidating the mechanisms underlying their action and importantly to design small molecule inhibitors through a structure-guided approach.
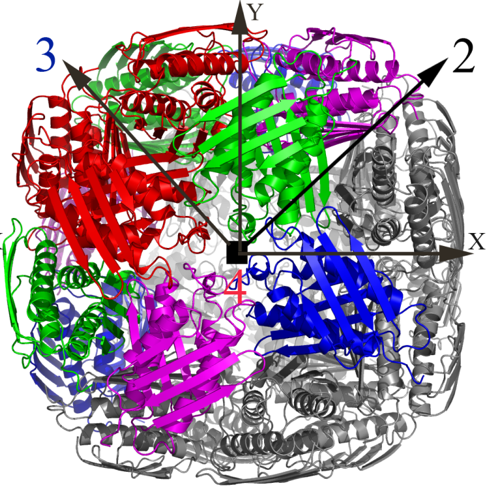
Over the past few years, we have determined high resolution crystal structures of imidazole glycerol phosphate dehydratase (HisB) and histidinol phosphate aminotransferases (HisC).HisB, which catalyses the conversion of imidazole glycerol phosphate to imidazole acetol phosphate (IAP), is comprised of 210 residues and folds as a single domain. The overall Mtb HisB structure resembles that of Arabidopsis thaliana, Filobasidiella neoformans, and Staphylococcus aureus. HisB is pseudo symmetric and is made up of a four-helix bundle sandwiched between two four-stranded mixed ß-sheets. The biological functional unit exhibits a quaternary assembly composed of 24 identical subunits with 432 molecular symmetry (Figures 2A & 2B) and possesses as many catalytic centres.
|
|
Figure 1. A schematic representation of the enzymatic steps that lead to the synthesis of His in M. tuberculosis. |
HisC catalyses the seventh step, the conversion of IAP to L-histidinol phosphate. It, like other PLP-dependent aminotransferases, consists of two domains, a larger PLP-binding domain having an α/ß/α topology and a smaller C-terminal domain. The spatial arrangement of the structural elements of HisC can be described as analogous to a curved left hand with three distinct clustering of structural motifs in the palm, thumb and fingers positions (Figure 3). The PLP-binding domain (palm) encompasses a bulk portion of the polypeptide chain and consists of a seven-stranded β-sheet sandwiched between six α- helices. The thumb domain, comprised of a bundle of three helices, serves as a junction connecting palm and finger domains. Protruded from this domain is an approximately 40-residue long loop, N-terminal arm, which closes over the PLP-binding domain (Figure 3). Recently, using a multidisciplinary approach we have shown that to counter a host directed histidine starvation, M. tuberculosis uses its de novo histidine pathway (Figure 4). This observation shows that chemical inhibition of this pathway may result in curtailing TB infection. In this respect, using structural and biochemical information, we have designed a number of triazole scaffold inhibitors for HisB. These compounds show sub-micromolar activity in vitro. Their in vivo efficacy is being examined.
|
|
|
Figure 2A. The X-ray structure of the biological unit of Mtb HisB in cartoon representation. The molecular symmetry 432 are shown by arrows. |
Figure 2B. Cryo-EM structure of Mtb HisB. |

|
 |
| Figure 3. Cartoon and surface representations of Mtb HisC. Shown in stick model are the bound PLP and Succinate. | Figure 4. A schematic representation depicting the IFN-γ-mediated host immune response and the counter response by Micobacterium Tuberculosis |
Mtb genome encodes approximately a dozen of membrane proteases (MPs). These molecules are important for Mtb virulence and survival in the adverse host macrophages. However, little is known with regard to their physiological substrates, disposition in the membrane, localization etc. We aim to address some of these questions mainly using biochemical, biophysical and immunological approaches. These require milligram quantities of monodisperse samples. We have over-expressed three recombinant version of MPs (Rv2223c, Rv2224c and Rv2672) in Mycobacterium smegmatis expression system and purified to a degree of homogeneity suitable for biophysical and biochemical studies. Recently we have shown that Rv2672 is involved in host lipids catabolism.
Deepsikha Kar, Soumya Banerjee, Swati Kumari, Varun Kumar, Mohini, Shivika Goyal, Yojana Vaidya, Vivek Kumar
Bacteria, plants and fungi all have enzymes to synthesize histidine (His). However, it cannot be synthesized de novo in humans due to the absence of equivalent enzymes. Mycobacterium tuberculosis (Mtb), the organism that causes tuberculosis (TB) in humans, makes His in 10 steps by employing 11 enzymes (Figure 1). Earlier studies by other groups and current data from my own group showed that even a single His pathway gene knockout significantly attenuates the bacterial load than the wild strain. In light of this result and the fact that His constitutes one of the building blocks of protein synthesis, inhibition of the enzymes that are involved in its biosynthesis gleams a rational strategy for new anti-TB agents design. In this context, the 3-D structures of these enzymes will be helpful in elucidating the mechanisms underlying their action and importantly to design small molecule inhibitors through a structure-guided approach.
Over the past few years, we have determined high resolution crystal structures of imidazole glycerol phosphate dehydratase (HisB) and histidinol phosphate aminotransferases (HisC).HisB, which catalyses the conversion of imidazole glycerol phosphate to imidazole acetol phosphate (IAP), is comprised of 210 residues and folds as a single domain. The overall Mtb HisB structure resembles that of Arabidopsis thaliana, Filobasidiella neoformans, and Staphylococcus aureus. HisB is pseudo symmetric and is made up of a four-helix bundle sandwiched between two four-stranded mixed ß-sheets. The biological functional unit exhibits a quaternary assembly composed of 24 identical subunits with 432 molecular symmetry (Figures 2A & 2B) and possesses as many catalytic centres.
 |
|
Figure 1. A schematic representation of the enzymatic steps that lead to the synthesis of His in M. tuberculosis. |
HisC catalyses the seventh step, the conversion of IAP to L-histidinol phosphate. It, like other PLP-dependent aminotransferases, consists of two domains, a larger PLP-binding domain having an α/ß/α topology and a smaller C-terminal domain. The spatial arrangement of the structural elements of HisC can be described as analogous to a curved left hand with three distinct clustering of structural motifs in the palm, thumb and fingers positions (Figure 3). The PLP-binding domain (palm) encompasses a bulk portion of the polypeptide chain and consists of a seven-stranded β-sheet sandwiched between six α- helices. The thumb domain, comprised of a bundle of three helices, serves as a junction connecting palm and finger domains. Protruded from this domain is an approximately 40-residue long loop, N-terminal arm, which closes over the PLP-binding domain (Figure 3). Recently, using a multidisciplinary approach we have shown that to counter a host directed histidine starvation, M. tuberculosis uses its de novo histidine pathway (Figure 4). This observation shows that chemical inhibition of this pathway may result in curtailing TB infection. In this respect, using structural and biochemical information, we have designed a number of triazole scaffold inhibitors for HisB. These compounds show sub-micromolar activity in vitro. Their in vivo efficacy is being examined.

|

|
|
Figure 2A. The X-ray structure of the biological unit of Mtb HisB in cartoon representation. The molecular symmetry 432 are shown by arrows. |
Figure 2B. Cryo-EM structure of Mtb HisB. |

|
 |
| Figure 3. Cartoon and surface representations of Mtb HisC. Shown in stick model are the bound PLP and Succinate. | Figure 4. A schematic representation depicting the IFN-γ-mediated host immune response and the counter response by Micobacterium Tuberculosis |
Mtb genome encodes approximately a dozen of membrane proteases (MPs). These molecules are important for Mtb virulence and survival in the adverse host macrophages. However, little is known with regard to their physiological substrates, disposition in the membrane, localization etc. We aim to address some of these questions mainly using biochemical, biophysical and immunological approaches. These require milligram quantities of monodisperse samples. We have over-expressed three recombinant version of MPs (Rv2223c, Rv2224c and Rv2672) in Mycobacterium smegmatis expression system and purified to a degree of homogeneity suitable for biophysical and biochemical studies. Recently we have shown that Rv2672 is involved in host lipids catabolism.



























